Compare Data from Different Sensitivity Experiments
Here, we develop a comparison between ICON data from experiments
Load Python Libraries
[1]:
%matplotlib inline
# system libs
import os, sys, glob
# array operators and netcdf datasets
import numpy as np
import xarray as xr
xr.set_options(keep_attrs=True)
import datetime
# plotting
import pylab as plt
import matplotlib.dates as mdates
myFmt = mdates.DateFormatter('%H:%M')
import seaborn as sns
sns.set_context('talk')
# drawing onto a map
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import cartopy.io.shapereader as shpreader
[2]:
sys.path.append( '/work/bb1224/2023_MS-COURSE/tools/analysis' )
from tools import convert_timevec
Open Data with xarray
General Paths
[3]:
exercise_path = '/work/bb1224/2023_MS-COURSE'
data_path = f'{exercise_path}/data'
icon_data_path = f'{data_path}/icon-lem'
This is the content of the data directory:
[4]:
os.listdir(icon_data_path)
[4]:
['grids-extpar', 'experiments', 'bc-init']
[5]:
grid_name = 'lpz_r2'
grid_path = f'{icon_data_path}/grids-extpar/{grid_name}'
[6]:
exp_list = ['base', 'sens01', 'sens02']
# exp_list = ['base', 'sens02']
exp_main_path = f'{icon_data_path}/experiments'
Grid File
[7]:
grid = xr.open_dataset( f'{grid_path}/lpz_r2_dom01_DOM01.nc', chunks={} )
Data Files
Now, we loop over different experiment paths.
[8]:
dlist = []
for exp_name in exp_list:
# set the file list
file_list = f'{exp_main_path}/icon_lam_1dom_lpz-{exp_name}/2d_cloud_DOM01_ML_20210516T*Z.nc'
# open data with xarray
dset = xr.open_mfdataset( file_list, chunks={'time':1}, combine='by_coords' )
dset['time'] = convert_timevec( dset.time.data )
# create a new experiment dimension
dset = dset.expand_dims( 'expname' )
dset['expname'] = [exp_name, ]
# collect all datasets in a list
dlist += [dset, ]
# combine the listed datasets again
dset = xr.concat( dlist, dim = 'expname' )
[9]:
d_mean = dset.mean( 'ncells' )
Prepare Precip Data
[10]:
rain_mean = d_mean['rain_con_rate'] + d_mean['rain_gsp_rate']
rain_mean.attrs['long_name'] = 'rain rate'
[11]:
conversion_factor = 3600 * 24
[12]:
rain_mean = conversion_factor * rain_mean
rain_mean.attrs['units'] = 'mm day-1'
[13]:
rain_mean = rain_mean.compute()
Plotting
[14]:
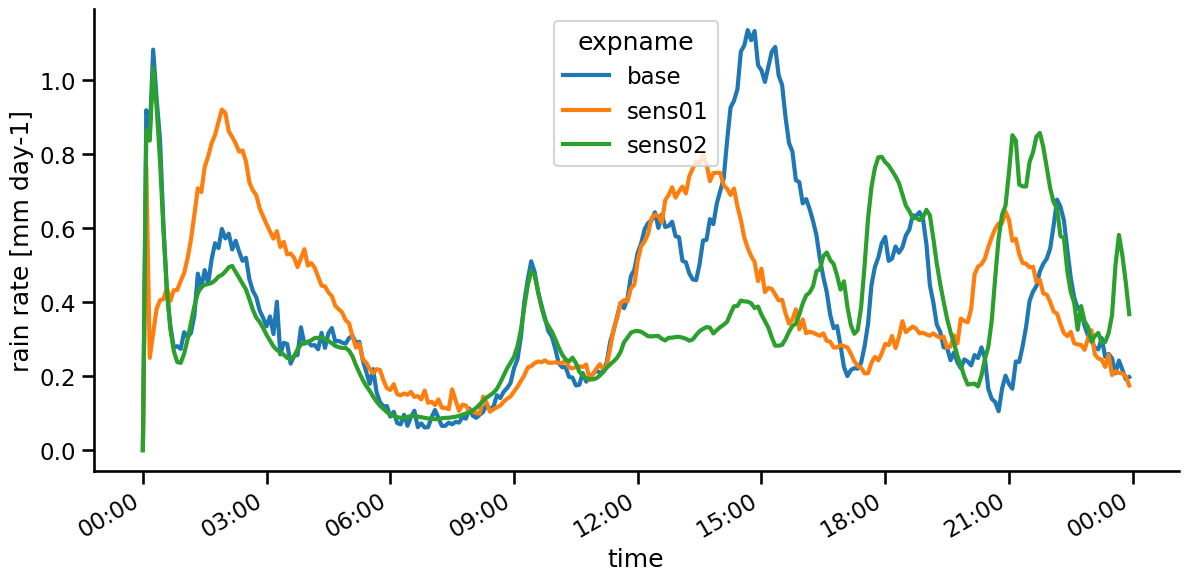
fig = plt.figure( figsize = (14,6))
plt.gca().xaxis.set_major_formatter(myFmt)
rain_mean.plot( hue = 'expname', lw = 3 )
sns.despine()
[15]:
rain_conv = d_mean['rain_con_rate']
rain_conv = conversion_factor * rain_conv
rain_conv.attrs['units'] = 'mm day-1'
rain_conv = rain_conv.compute()
[16]:
rain_gridscale = d_mean['rain_gsp_rate']
rain_gridscale = conversion_factor * rain_gridscale
rain_gridscale.attrs['units'] = 'mm day-1'
rain_gridscale = rain_gridscale.compute()
[17]:
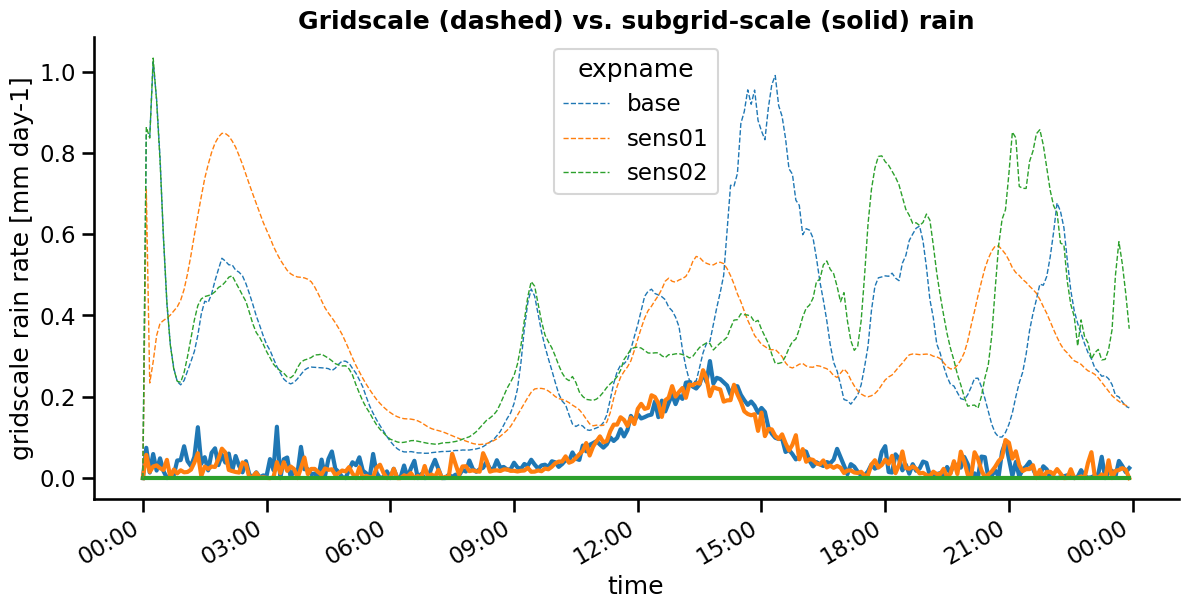
fig = plt.figure( figsize = (14,6))
plt.gca().xaxis.set_major_formatter(myFmt)
rain_conv.plot( hue = 'expname', lw = 3,)
plt.gca().set_prop_cycle(None)
rain_gridscale.plot( hue = 'expname', ls = '--', lw = 1 )
sns.despine()
plt.title( 'Gridscale (dashed) vs. subgrid-scale (solid) rain', fontweight = 'bold')
[17]:
Text(0.5, 1.0, 'Gridscale (dashed) vs. subgrid-scale (solid) rain')
[18]:
rain_mean.mean('time')
[18]:
<xarray.DataArray (expname: 3)>
array([0.48379675, 0.39261702, 0.47736754])
Coordinates:
* expname (expname) <U6 'base' 'sens01' 'sens02'
Attributes:
standard_name: rain_con_rate
long_name: rain rate
units: mm day-1
param: 76.1.0
CDI_grid_type: unstructured
number_of_grid_in_reference: 1Tasks
Think about the question:
Is convective rain changed when gridscale microphysics is switched from two-moment to one-moment bulk scheme?
If not, why?
Please explore other data with this notebook! Check how the temporal evolution looks like for
total cloud cover, variable name
clctliquid water path, variable name:
tqc_diaice water path, variable name:
tqi_dia